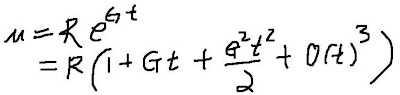# ~/0.network.aging.prj/0a.network.fitting/_binomial_aging20140711.r
#test binomial aging model
rm(list=ls());
source("lifespan.20131221.r")
require(flexsurv)
##### log likelihood function, 3-parameter binomial mortality rate model
# http://hongqinlab.blogspot.com/2013/12/binomial-mortailty-model.html
# m = R ( 1 + t/t0)^(n-1)
# s = exp( (R t0/n)*(1 - (1+t/t0)^n ) )
llh.binomialMortality.single.run <- function( Rt0n, lifespan ) {
I = Rt0n[1]; t0 = Rt0n[2]; n=Rt0n[3];
my.data = lifespan[!is.na(lifespan)];
log_s = (I * t0 /n )*(1 - (1 + my.data/t0)^n);
log_m = log(I) + (n-1) * log(1 + my.data/t0 );
my.lh = sum(log_s) + sum(log_m);
print (Rt0n ); #trace the convergence
ret = - my.lh # because optim seems to maximize
}
#random number from binomial-aging model
rbinomial.aging <- function ( Rt0n, n){
x.uniform = runif(n)
inverse.binomialaging.CDF = function(R,t0,n,y) {t0*((1- n*log(y)/(R*t0))^(1/n) -1) }
x.binomialaging = inverse.binomialaging.CDF(Rt0n[1],Rt0n[2], Rt0n[3], x.uniform)
return(x.binomialaging)
}
set.seed(20140711)
x = rbinomial.aging( c(0.001,10, 5), 1000)
fitBinom = optim ( c(0.001,10, 3), llh.binomialMortality.single.run , lifespan=x,
method="L-BFGS-B", lower=c(1E-10,0.05,1E-1), upper=c(10,100,50) );
fitGomp = optim ( c(0.005, 0.08), llh.G.single.run , lifespan=x,
method="L-BFGS-B", lower=c(1E-10,1E-2), upper=c(10,10) );
fitWeib2 = flexsurvreg(formula = Surv(x) ~ 1, dist="weibull")
fitGomp2 = flexsurvreg(formula = Surv(x) ~ 1, dist="gompertz")
fitGomp2$coeff
c(-fitGomp$value, fitGomp2$loglik, fitWeib2$loglik, -fitBinom$value)
Flexsurv can over-fit the Gompertz with negative values, and Weibull gave the best value!!!
Even though the random number are generated by binomial aging model, its fitting gave the lowest likelihood?!
TODO: I should to provide a gradient function for optim(method='L-BFGS-B')
Or try optimx()
For binomial-aging, $n$ is under-estimated by optim(), but $m0$ is over-estimated. When the entire data is fitted with Gompertz model, $G$ is too low. How about fitting within initial stage?
set.seed(20140711)
x = round( rbinomial.aging( c(0.001,10, 4), 5000), digits=1 )#t0=10, n=4
tb = calculate.mortality.rate(x)
plot( tb$s ~ tb$t)
plot( tb$mortality.rate ~ tb$t )
plot( tb$mortality.rate ~ tb$t, log='y', main='linear for Gompertz' )
plot( tb$mortality.rate ~ tb$t, log='yx', main='linear for Weibull' )
set.seed(20140711)
x = rbinomial.aging( c(0.01,25, 4), 100) #large m0, t0=25, n=4
x = round( x + 0.5, digits=0)
fitBinom = optim ( c(0.01,25, 3.5), llh.binomialMortality.single.run , lifespan=x,
method="L-BFGS-B", lower=c(1E-10,0.05,1E-1), upper=c(10,100,50) );
fitGomp = optim ( c(0.005, 0.08), llh.G.single.run , lifespan=x,
method="L-BFGS-B", lower=c(1E-10,1E-2), upper=c(10,10) );
fitWeib2 = flexsurvreg(formula = Surv(x) ~ 1, dist="weibull")
fitGomp2 = flexsurvreg(formula = Surv(x) ~ 1, dist="gompertz")
fitGomp2$coeff
c(-fitGomp$value, fitGomp2$loglik, fitWeib2$loglik, -fitBinom$value)
Good, binomial aging model gives the best likelihood.
Plot confirmed that Gompertz looks better than Weibull.