=> pentagon glucose on polarity of DNA strand. Glucose said, "I am the decider."
=> electron transport chain, dam, and power
data analysis cartoon
http://stats.stackexchange.com/questions/423/what-is-your-favorite-data-analysis-cartoon
This site is to serve as my note-book and to effectively communicate with my students and collaborators. Every now and then, a blog may be of interest to other researchers or teachers. Views in this blog are my own. All rights of research results and findings on this blog are reserved. See also http://youtube.com/c/hongqin @hongqin
Tuesday, September 30, 2014
BIO233 20140930 Ch5 continued, and Ch6 molecular biology
Socrative setup, Room: HongQin
Plating question from last week
SPDBViewer on 5' and 3' ends of DNA double helix (only 1 student identify the 3'end)
paper and razor, paper sequence for PCR reactions
Use yeast COX1 coding sequence (this mito gene contains introns)
http://www.yeastgenome.org/locus/S000007260/sequence
The sequences are printed too close to each other. I did not print enough primers. I did not provide scotch tapes for students to finish the reaction.
x Exercises were given back to students when they were working on problem sets.
x Exams back to students
Monday, September 29, 2014
reverse complementary sequences in R seqinr
##
## Show that comp() does *not* return the reverve complementary strand:
##
c2s(comp(s2c("aaaattttggggcccc")))
##
## Show how to get the reverse complementary strand:
##
c2s(rev(comp(s2c("aaaattttggggcccc"))))
##
## Show what happens with non allowed values:
##
c2s(rev(comp(s2c("aaaaXttttYggggZcccc"))))
##
## Show what happens with ambiguous bases:
##
allbases <- s2c("abcdghkmstvwn")
comp(allbases) # NA are produced
comp(allbases, ambiguous = TRUE) # No more NA
my code: _get_double_strand.R
setwd("~/github/ctls/sequences")
install.packages("seqinr")
library(seqinr);
list.files()
seqs = read.fasta("panda16srDNA.fasta")
str(seqs);
seqs[[1]]
length(seqs[[1]])
c2s( rev( comp(seqs[[1]]) ) )
c2s(seqs[[1]][1:120])
c2s(comp(seqs[[1]][1:120]))
Saturday, September 27, 2014
LARPS, PNAS 2014
Protein mass spec data is deposited, and can be used to estimate protein half-life with respect to RLS? .... .... I looked at the deposited data, but there is no meta-data.
The author use ~135 because they did not over-extend themselves. Their arbitrary cutoff give 135 nuclear genes and 1 mitochondrial gene COX1.
Does LARPs tend to have few carbonylation motif?
Does LARPs tend to have smaller lambda for gene interaction in my network aging model?
The author use ~135 because they did not over-extend themselves. Their arbitrary cutoff give 135 nuclear genes and 1 mitochondrial gene COX1.
Does LARPs tend to have few carbonylation motif?
Does LARPs tend to have smaller lambda for gene interaction in my network aging model?
Negative correlation between PlasmidLoss measure and initial LOH mitotic asymmetry measure (L0)
With only 6 strains, there is a negative correlation between mitotic asymmetry (L0) and first plasmid loss measure, pvalue = 0.04. (I missed M1-2. It seems to be labeled as M12 in the plasmid loss data set. After add M1-2, partial correlation L0.small ~ PL1 + CLS still works.
The second plasmid loss measure is not significantly correlated.
#> pTb[pTb<0.05] #PL2
#<NA> Nloh <NA> PL1 PL2 PL
#NA 0.0111638902 NA 0.0022934461 0.0000000000 0.0001032832
#> pTb[pTb<0.09] #PL1
#<NA> Nloh b.min.sd TLmax.sd <NA> L0.small PL1 PL2 PL
#NA 3.138762e-02 8.707534e-02 5.595864e-02 NA 3.957404e-02 0.000000e+00 2.293446e-03 5.375196e-05
The combined measure has a pvalue = 0.06.
#> pTb[pTb<0.09]
#<NA> Nloh b.min.sd TLmax.sd <NA> L0.small PL1 PL2 PL
#NA 1.647319e-02 5.877810e-02 7.604223e-02 NA 6.414451e-02 5.375196e-05 1.032832e-04 0.000000e+00
Partial correlation tb$L0.small ~ tb$PL1 + tb$CLS is still good. (but not with PL2)
TODO: H2O2 data, Tg/Tc
See: github/0.network.aging.prj/10.choy/_choy.plasmidLoss.20140926.R
rm(list=ls())
require(xlsx)
setwd("~/projects/0.network.aging.prj/10.choy")
list.files()
plasmidlosstb = read.xlsx("plasmid loss assays.xlsx", 1)
names(plasmidlosstb)= c("strain", "PL1", "PL2")
plasmidlosstb$strain = as.character( plasmidlosstb$strain )
str(plasmidlosstb)
LOHtb = read.table("021307.summary.by.strain.csv", sep="\t", header=T)
LOHtb$strain = as.character(LOHtb$strain)
LOHtb$PL1 = NA; LOHtb$PL2 = NA; LOHtb$PL=NA;
str(LOHtb[,1:10])
for( i in 1:length(LOHtb$strain) ){
mys = LOHtb$strain[i]
# mys ="M8"; #debug
x = grep(mys, plasmidlosstb$strain)
if( length(x)>0 ) {
tmptb = plasmidlosstb[x, ]
LOHtb$PL1[i] = mean(tmptb$PL1, na.rm=T)
LOHtb$PL2[i] = mean(tmptb$PL2, na.rm=T)
LOHtb$PL[i] = mean( c(tmptb$PL1, tmptb$PL2), na.rm=T )
}else {
print(paste(mys, " is not found in plasmid loss data"))
}
}
### all possible regression analysis for a given column
tb = LOHtb
pTb = 1: length(tb[1,])
names(pTb) = names(tb)
for( j in c(2:40) ) {
m = lm( tb[, j] ~ tb$PL)
#m = lm( tb[, j] ~ tb$PL1)
#m = lm( tb[, j] ~ tb$PL2)
sm = summary(m)
pTb[j] = 1 - pf(sm$fsta[1], sm$fsta[2], sm$fsta[3])
}
pTb[pTb<0.09]
#> pTb[pTb<0.05] #PL2
#<NA> Nloh <NA> PL1 PL2 PL
#NA 0.0111638902 NA 0.0022934461 0.0000000000 0.0001032832
#> pTb[pTb<0.09] #PL1
#<NA> Nloh b.min.sd TLmax.sd <NA> L0.small PL1 PL2 PL
#NA 3.138762e-02 8.707534e-02 5.595864e-02 NA 3.957404e-02 0.000000e+00 2.293446e-03 5.375196e-05
#> pTb[pTb<0.09]
#<NA> Nloh b.min.sd TLmax.sd <NA> L0.small PL1 PL2 PL
#NA 1.647319e-02 5.877810e-02 7.604223e-02 NA 6.414451e-02 5.375196e-05 1.032832e-04 0.000000e+00
m = lm(tb$L0.small ~ tb$PL1)
summary(m)
plot( tb$L0.small ~ tb$PL1, pch=18, xlim=c(2,7), ylim=c(0.05, 0.20))
abline(m, col='red')
text( tb$PL1, tb$L0.small, tb$strain)
text( 3, 0.1, "R2=0.69, p=0.04")
Labels:
aging,
collaboration,
LOH,
plasmid loss,
yeast,
yeast aging
Friday, September 26, 2014
mitochondrial gene networks (need more information
gene expression networks
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0057739
(essential genes can be identified by orthologs of E coli and Sce genes)
Thursday, September 25, 2014
Spelman Grade percentage score to letter grade
Grade scale, grade distribution.
Grade Scale for letter grade
Grade Percentage Score
A 94
- 100
A- 90 - 93.9
B+ 87 – 89.9
B 84 – 86.9
B- 80 – 83.9
C+ 76 – 79.9
C 70 – 75.9
C- 67 – 69.9
D+ 64 – 66.9
D 60 – 63.9
F 0 - 59.9
grade2letter = function(x){
if(x>94){ ret='A'
}else if (x >90) { ret='A-'
}else if (x >87 ){ ret = 'B+'
}else if (x > 84){ ret = 'B'
}else if (x >80){ ret = 'B-'
}else if (x > 76){ ret = 'C+'
}else if (x > 70){ ret = 'C'
}else if (x > 67){ ret = 'C-'
}else if (x > 64){ ret = 'D+'
}else if (x > 60){ ret = 'D'
}else { ret = 'F'
}
return (ret)
}
grade2letter = function(x){
if(x>94){ ret='A'
}else if (x >90) { ret='A-'
}else if (x >87 ){ ret = 'B+'
}else if (x > 84){ ret = 'B'
}else if (x >80){ ret = 'B-'
}else if (x > 76){ ret = 'C+'
}else if (x > 70){ ret = 'C'
}else if (x > 67){ ret = 'C-'
}else if (x > 64){ ret = 'D+'
}else if (x > 60){ ret = 'D'
}else { ret = 'F'
}
return (ret)
}
bio233 chapter 5, microbial growth, hemocytometer, standard deviation
30 minutes Worked on hemocytometer usage. I put 73 dots on a hemocytometer grid print-out.
40 minutes Worked on a GRE microbial temperature problem
30 minutes, Worked on a dilution questions again. (did not get into the plating question).
To explain standard deviation, I drew a chart on the board, with average as the horizontal line, and dots above or below the line. Squares are then drawn to explains the variance and standard deviations.
I was not clear on the interpret of standard deviation. One stddev should be 68%, two stddev should be 95%.
The concepts that students found difficult are:
1. How the volume of hemocytometer square is decided.
Some students did
2. unit conversion from ul^-1 to ml^-1 during concentration calculation. When the unit is in the denominator, it seems to throw some student off balance.
3. Standard deviation
Wednesday, September 24, 2014
streak met15+/-s strains
The strains mostly at the bottom of their vials due to freezer breakdown one time.
So, I let the vials thaw at RT from 4:21pm-4:30pm.
The vials were completely thawed after 10 minutes at room temperature.
I used suspended the cells with filtered tips, and put them a new MLA plate.
previous plate
I put strains on MLA plates. Some strains grew to tiny colonies in 1 day. Others took 4-5 days to form visible colonies.
So, I let the vials thaw at RT from 4:21pm-4:30pm.
The vials were completely thawed after 10 minutes at room temperature.
I used suspended the cells with filtered tips, and put them a new MLA plate.
previous plate
I put strains on MLA plates. Some strains grew to tiny colonies in 1 day. Others took 4-5 days to form visible colonies.
LOH Pbt0 L0 in natural isolates
So, YPS128 and 163 have higher L0.
Text format:
| Lmax.sd | strains | L0.all | L0.all/min | Pb0/minPb0 | Pbt0 | strain |
| 0.46808119 | 101S | 0.15957447 | 2.746360404 | 1.522807342 | 0.00280093 | 101S |
| 0.31005376 | M1-2 | 0.06164384 | 1.060922849 | 1 | 0.00183932 | M1-2 |
| 0.44523402 | M13 | 0.09345794 | 1.608460212 | 1.07483744 | 0.00197697 | M13 |
| 0.88881944 | M14 | 0.1122449 | 1.931793657 | 3.804221125 | 0.00699718 | M14 |
| 2.84785884 | M2-8 | 0.14859438 | 2.557387291 | 1.992959355 | 0.00366569 | M2-8 |
| 0.08020806 | M32 | 0.10962567 | 1.886715333 | 6.716993237 | 0.0123547 | M32 |
| 0.35994212 | M34 | 0.17134831 | 2.948994372 | 3.952901072 | 0.00727065 | M34 |
| 1.26301227 | M5 | 0.171875 | 2.958058983 | 1.090267055 | 0.00200535 | M5 |
| 0.96576395 | M8 | 0.05810398 | 1 | 3.464068243 | 0.00637153 | M8 |
| 1.33810812 | YPS128 | 0.18353175 | 3.15867777 | 5.90821608 | 0.0108671 | YPS128 |
| 0.98348022 | YPS163 | 0.25268817 | 4.348896065 | 3.872534415 | 0.00712283 | YPS163 |
| NA | BY4743 | 0.18918919 | 3.256045283 | 3.176902333 | 0.00584334 | BY4743 |
E coli genomics resources
Strains
E. coli Genetic Resources at Yale
http://cgsc.biology.yale.edu/KeioList.php
E coli PPI
CGSC
Coli Genetic Stock Center
Coli Genetic Stock Center
E. coli Genetic Resources at Yale
CGSC, The Coli Genetic Stock Center
http://cgsc.biology.yale.edu/KeioList.phpE coli PPI
coli k-12, Genome Res., 2006, 16(5), 686–91.
Essential gene interactions
https://www.jstage.jst.go.jp/article/ggs/88/4/88_233/_article
E coli essential genes
http://www.shigen.nig.ac.jp/ecoli/pec/ Profiles of Escherichia coli chromosomes (PEC)
302 essential genes, 4400 nonessential genes
Used in Jordan02GR paper
303 essential genes in E coli K12, Ecogene
http://www.ecogene.org/old/topic.php?topic_id=5
295 essential genes in Ecowiki
http://ecoliwiki.net/colipedia/index.php/Essential_genes
Gerdes03JB, essential genes in E coli MG1655
http://jb.asm.org/content/185/19/5673.short
620 essential gene, 3126 non-essential genes
http://www.genome.wisc.edu/resources/essential.htm
Essential genome of Caulobacter crescentus
http://onlinelibrary.wiley.com/doi/10.1038/msb.2011.58/full
Homologs of B subtilis essential genes?
EcoK12 Expression data, GO
http://www.porteco.org/downloads/dataAccess/
Essential gene interactions
https://www.jstage.jst.go.jp/article/ggs/88/4/88_233/_article
E coli essential genes
http://www.shigen.nig.ac.jp/ecoli/pec/ Profiles of Escherichia coli chromosomes (PEC)
302 essential genes, 4400 nonessential genes
Used in Jordan02GR paper
303 essential genes in E coli K12, Ecogene
http://www.ecogene.org/old/topic.php?topic_id=5
295 essential genes in Ecowiki
http://ecoliwiki.net/colipedia/index.php/Essential_genes
Gerdes03JB, essential genes in E coli MG1655
http://jb.asm.org/content/185/19/5673.short
620 essential gene, 3126 non-essential genes
http://www.genome.wisc.edu/resources/essential.htm
Essential genome of Caulobacter crescentus
http://onlinelibrary.wiley.com/doi/10.1038/msb.2011.58/full
Homologs of B subtilis essential genes?
EcoK12 Expression data, GO
http://www.porteco.org/downloads/dataAccess/
Tuesday, September 23, 2014
bio233 exam1, part2, openbook
I asked even number students to start the computer-based questions, and odd number to begin with paper-based questions.
Wireless connection problem occurred to 4-5 students. I tried scwifi, and thad to use Ethernet cable.
Ten HP laptops missing power cords.
I forgot to set time limit on the online open-book test, and some students very long time.
I forgot the change the [substrate] label for a plot. In the surface/volume questions, it should be diameter not radius.
One student mistook "coefficient of determination" (R-squared) as "correlation coefficient" (r).
I was busy with the wireless problem for about 40-50 minutes.
Wireless connection problem occurred to 4-5 students. I tried scwifi, and thad to use Ethernet cable.
Ten HP laptops missing power cords.
I forgot to set time limit on the online open-book test, and some students very long time.
I forgot the change the [substrate] label for a plot. In the surface/volume questions, it should be diameter not radius.
One student mistook "coefficient of determination" (R-squared) as "correlation coefficient" (r).
I was busy with the wireless problem for about 40-50 minutes.
SVM, multiple categories, TOREAD
Common vaccinations
DT Diphtheria 白喉 and tetanus
toxoids
DTaP (is similar but with “a”
for acelluar”).
OPV (oral polio vaccine)
IPV (inactivated polio vaccine)
MMR(Measles, Mumps, Rubella)
麻腮风三联疫苗
Monday, September 22, 2014
bio233 Gram stain 20140921 Monday
2pm-4:30pm.
I played previous youtube demo, but the angle did not show details. I then did a demo in class.
I forgot to use 70% ethanol to clean my workspace for the demo.
Some students flamed the loops with the bottom of the flame.
Some students tried multiple times to get the staining working. These are good students that really want to learn.
I asked the students to clean up after the lab.
E coli from Carolina biology has a cocci shape. Bacillus cereus shipped is also contaminated with cocci bacteria.
I played previous youtube demo, but the angle did not show details. I then did a demo in class.
I forgot to use 70% ethanol to clean my workspace for the demo.
Some students flamed the loops with the bottom of the flame.
Some students tried multiple times to get the staining working. These are good students that really want to learn.
I asked the students to clean up after the lab.
E coli from Carolina biology has a cocci shape. Bacillus cereus shipped is also contaminated with cocci bacteria.
Friday, September 19, 2014
bio386 learning R using the metric example
Let the student took the online metric survey.
I then worked with the students from line 1-100 in the metric_v2.R
We went through looking for helps, object, $ and indexing, loops, parentheses
read.csv
table,
cleaning data
An error was found in the tb2$age calculation.
At the end of class, I asked the student to sign up the Coursera "R programming" course.
I then worked with the students from line 1-100 in the metric_v2.R
We went through looking for helps, object, $ and indexing, loops, parentheses
read.csv
table,
cleaning data
An error was found in the tb2$age calculation.
At the end of class, I asked the student to sign up the Coursera "R programming" course.
Thursday, September 18, 2014
FYE, how to study 20140918 Thu
http://www.howtostudy.com/
Notes, emory visit 20140918
For t-test, if samples are normally distributed, t-test is accurate even for small sample size. For large sample size, because of the central limit theorem, t-test is also accurate. This is the reason t-test is a 'robust' method. According to John, there is not general theory on why a method is robust or not, because it depend on how the method will be used. John said that reliability is in the field of operational research.
Tianwei suggest that binned sample may not be uniformally distributed, so a taking the center may add a 'bias' to the rounding procedure.
For the Emory biostat program, GRE verbal is required over 50%, and quantitative part is over 80%. Their experience is that GRE score is the best predictor of student performance in the program, especially the quantitative score.
Tianwei suggest that binned sample may not be uniformally distributed, so a taking the center may add a 'bias' to the rounding procedure.
For the Emory biostat program, GRE verbal is required over 50%, and quantitative part is over 80%. Their experience is that GRE score is the best predictor of student performance in the program, especially the quantitative score.
Wednesday, September 17, 2014
FYE, Personal statement, samples, Medical school application
Bad example, too general
http://www.accepted.com/medical/sampessay09.aspx
As the time approached for me to set my personal and professional goals, I made a conscientious decision to enter a field which would provide me with a sense of achievement and, at the same time, produce a positive impact on mankind. It became apparent to me that the practice of medicine would fulfill these objectives. In retrospect, my ever-growing commitment to medicine has been crystallizing for years. My intense interest in social issues, education, and athletics seems particularly appropriate to this field and has prepared me well for such a critical choice...
I’ve been asked many times why I wish to become a physician. Upon considerable reflection, the thought of possessing the ability to help others provides me with tremendous internal gratification and offers the feeling that my life’s efforts have been focused in a positive direction. Becoming a physician is the culmination of a lifelong dream; and I am prepared to dedicate myself, as I have in the past, to achieving this goal.
The revised one:
Crayfish tails in tarragon butter, galantine of rabbit with foie gras, oxtail in red wine, and apple tartelettes. The patient had this rich meal and complained of "liver upset" (crise de foie). Why a liver ache? I always associate indigestion with a stomach ache. In studying French culture in my Evolutionary Psychology class, I learned that when experiencing discomfort after a rich meal, the French assume their liver is the culprit. Understanding and dealing with the minor — sometimes major — cultural differences is a necessity in our shrinking world and diverse American society. Anthropology has prepared me to effectively communicate with an ethnically diverse population. My science classes, research, and clinical experience have prepared me to meet the demands of medical school.
I first became aware of the valuable service that physicians provide when I observed my father, a surgeon, working in his office. I gained practical experience assisting him and his staff perform various procedures in his out-patient center. This exposure increased my admiration for the restorative, technological, and artistic aspects of surgery. I also saw that the application of medical knowledge was most effective when combined with compassion and empathy from the health care provider.
While admiring my father's role as a head and neck surgeon helping people after severe accidents, I also found a way to help those suffering from debilitating ailments. Working as a certified physical trainer, I became aware of the powerful recuperative effects of exercise. I was able to apply this knowledge in the case of Sharon, a forty-three-year-old client suffering from lupus. She reported a 200% increase in her strength tests after I trained her. This meant she could once again perform simple tasks like carrying groceries into her house. Unfortunately, this glimpse of improvement was followed by a further deterioration in her condition. On one occasion, she broke down and cried about her declining health and growing fears. It was then that I learned no physical prowess or application of kinesiology would alleviate her pain. I helped reduce her anxiety with a comforting embrace. Compassion and understanding were the only remedies available, temporary though they were.
To confirm that medicine is the best way for me to help others, I assisted a research team in the Emergency Room at University Medical Center (UMC). This experience brought me in direct contact with clinical care and provided me with the opportunity to witness and participate in the "behind-the-scenes" hospital operations. Specifically, we analyzed the therapeutic effects of two new drugs — Drug A and Drug B — in patients suffering from acute ischemic stroke. The purpose of this trial was to determine the efficacy and safety of these agents in improving functional outcome in patients who had sustained an acute cerebral infarction. My duties centered around the role of patient-physician liaison, determining patients' eligibility, monitoring their conditions, and conducting patient histories.
I continued to advance my research experience at the VA Non-Human Primate Center. During the past year, I have been conducting independent research in endocrinology and biological aspects of anthropology. For this project, I am examining the correlation between captive vervet monkeys' adrenal and androgen levels with age, gender, and various behavioral measures across different stress-level environments. I enjoy the discipline and responsibility which research requires, and I hope to incorporate it into my career.
Anthropology is the study of humans; medicine is the science and art dealing with the maintenance of health and the prevention, alleviation, or cure of disease in humans. From my work at UMC and my observation of my father’s practice, I know medicine will allow me to pursue an art and science that is tremendously gratifying and contributes to the welfare of those around me. My anthropology classes have taught me to appreciate cross-cultural perspectives and their relationship to pathology and its etiology. First hand experience with exercise therapy and nutrition has taught me the invaluable role of prevention. Medical school will now provide me with the technical knowledge to alleviate a crise de foie.
http://prezi.com/rtqxofl_vpx5/examples-of-a-good-and-bad-argumentative-essay/
http://kimberlychapman.com/essay/badwriting.html
Maloney Lab meeting, 20140917
Maloney lab meeting
Glycolipids between VT500 and Daudi cell lines
GC
LC
Gb3 (aka CD77), a rare CD## is actually glycolipid
Gb4
Lipid raft in cell membrane contain high concentration of glycolipids
Lipid raft research bridges glycolipid and signal transduction
Gb3/CD77 = globotraisocyla ceramide = glycosphinglolipid
Defines pk blood group antigen
Receptor for Shiga toxin/verotoxins
Burkitt's lympphoma associated antigen
CD77 is a marker for germinal center B cells
Shiga toxin targets Gb3, arguing that some Gb3 should also bind some normal human proteins.
Shigtoxin is also called Verotoxin in Canada.
Gb3 may be invovled in cell-cell interaction
Bacterial cells can shutdown germinal cells and eliminate immune response memory by targeting Gb3.
Gb3 is involved in apoptosis signaling
Shiga toxin binding of Gb3 induces apoptosis, and this can be triggered with B-subunit alone.
---------
Burkitt's Lymphoma is a maligant lymphomas of African children.
Associated with malaria zones and EbV-infection
------
Germinal center cells
Antigen recognition by surface antibody (CD19 part of co-receptor complex).
Antigen presentation to CD4+T helper cells
Affinity maturation of antibody responses/Somatic hypermutations
Isotype switching
-----
Role of CD77 in Germinal center B cells
CD19 in one cell binds Gb3 on another cells. CD19 is a glyco-protein with a large extra-cellular protein domain.
CD19/CD77 binding as adhesion mechanism
MHC classII expression
-----
Interferon-alpha receptor subunit 1 contains a Shiga toxin like amino acid sequence.
CD19 also has a Shiga toxin like amino acid sequence. This sequence is distributed in 3 regions, with quite some distance between them.
-----
VT500
Daudi
How are they different in Gb3?
-----
apoptosis inducers: capersysin(?),
JC-1 dye to monitor mitochondrial membrane polarization
-----
Search of Shiga-toxin like sequence pattern can be improved by HMM.
Shiga-toxin is also shared by Shegalla boydii
Todo: orthologs of CD19 in other mamals, and their shiga toxin motifs.
It turns out that Shiga toxin does affect germ-line B cells in mouse, so mouse CD19 is not expected to has shiga-toxin binding motif. Well, how about Neanderthal?
Glycolipids between VT500 and Daudi cell lines
GC
LC
Gb3 (aka CD77), a rare CD## is actually glycolipid
Gb4
Lipid raft in cell membrane contain high concentration of glycolipids
Lipid raft research bridges glycolipid and signal transduction
Gb3/CD77 = globotraisocyla ceramide = glycosphinglolipid
Defines pk blood group antigen
Receptor for Shiga toxin/verotoxins
Burkitt's lympphoma associated antigen
CD77 is a marker for germinal center B cells
Shiga toxin targets Gb3, arguing that some Gb3 should also bind some normal human proteins.
Shigtoxin is also called Verotoxin in Canada.
Gb3 may be invovled in cell-cell interaction
Bacterial cells can shutdown germinal cells and eliminate immune response memory by targeting Gb3.
Gb3 is involved in apoptosis signaling
Shiga toxin binding of Gb3 induces apoptosis, and this can be triggered with B-subunit alone.
---------
Burkitt's Lymphoma is a maligant lymphomas of African children.
Associated with malaria zones and EbV-infection
------
Germinal center cells
Antigen recognition by surface antibody (CD19 part of co-receptor complex).
Antigen presentation to CD4+T helper cells
Affinity maturation of antibody responses/Somatic hypermutations
Isotype switching
-----
Role of CD77 in Germinal center B cells
CD19 in one cell binds Gb3 on another cells. CD19 is a glyco-protein with a large extra-cellular protein domain.
CD19/CD77 binding as adhesion mechanism
MHC classII expression
-----
Interferon-alpha receptor subunit 1 contains a Shiga toxin like amino acid sequence.
CD19 also has a Shiga toxin like amino acid sequence. This sequence is distributed in 3 regions, with quite some distance between them.
-----
VT500
Daudi
How are they different in Gb3?
-----
apoptosis inducers: capersysin(?),
JC-1 dye to monitor mitochondrial membrane polarization
-----
Search of Shiga-toxin like sequence pattern can be improved by HMM.
Shiga-toxin is also shared by Shegalla boydii
Todo: orthologs of CD19 in other mamals, and their shiga toxin motifs.
It turns out that Shiga toxin does affect germ-line B cells in mouse, so mouse CD19 is not expected to has shiga-toxin binding motif. Well, how about Neanderthal?
Spelman van, 15 passenger
The Physical Education Department has contracted for an instructor to conduct a Van Orientation Training
This training is required for anyone who wishes to operate a College owned or leased 15 passenger van.
Steve Bowser
Spelman College
Dir. Public Safety
(404) 270-5326
This training is required for anyone who wishes to operate a College owned or leased 15 passenger van.
Steve Bowser
Spelman College
Dir. Public Safety
(404) 270-5326
bio386 20140917 Wed
UTK meeting registration
Hotel verification
HHMI Letter of intent. Student read out loud and revised the letter.
Hotel verification
HHMI Letter of intent. Student read out loud and revised the letter.
Tuesday, September 16, 2014
Network resources, human, (in progress)
*** GeneSigDB (used by Li14BMC, should contain disease genes). This site provide download for R analysis. http://compbio.dfci.harvard.edu/genesigdb/
List of human disease genes in gene Card
http://www.genecards.org/cgi-bin/listdiseasecards.pl?type=full
(not sure whether this includes haploid type association)
Farmington study
http://videocast.nih.gov/Summary.asp?File=18760&bhcp=1
Network analysis of GWAS data, 2013 Current Opinion in Genetics and Development
MIF Parsers
The human protein interaction network data can be found from http://www.hprd.org/download
Reference: Peri S, Navarro JD, Amanchy R,
Kristiansen TZ, Jonnalagadda CK,
Surendranath V, Niranjan V, Muthusamy B, Gandhi TKB, GronborgM,
Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP,
Ramya MA, Zhao ZX, Chandrika KN, Padma N, Harsha HC et al
(2003) Development of human protein reference database as an initial
platform for approaching systems biology in humans. Genome Res 13:
2363–237
Gerstein lab's human networks
human multinet
http://homes.gersteinlab.org/Khurana-PLoSCompBio-2013/
http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1002886#s4
http://encodenets.gersteinlab.org/
dbCline
UChicago
peptieAtlas
http://www.nature.com/embor/journal/v9/n5/pdf/embor200856.pdf
ensemble
human twin aging expression
http://genomebiology.com/2013/14/7/R75?utm_campaign=10_12_13_genomebiol_Article_Mailing_Reg&utm_content=7387379393&utm_medium=BMCemail&utm_source=Emailvision
http://hongqinlab.blogspot.com/2013/10/mutation-tolerance-in-human-genes-data.html
http://hongqinlab.blogspot.com/2013/03/candidate-rojects-for-r-based-data.html
expression, aging, human, kidny,
http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0020427
From Gilman 2011 Neuron, netbag on autism
gene expression between human and mouse, PNAS, mike snyder lab, Standford
http://www.pnas.org/content/111/48/17224.full
List of human disease genes in gene Card
http://www.genecards.org/cgi-bin/listdiseasecards.pl?type=full
(not sure whether this includes haploid type association)
Genome Research Genome-wide map of regulatory interactions in the human genome,
http://genome.cshlp.org/content/early/2014/09/15/gr.176586.114.abstractFarmington study
http://videocast.nih.gov/Summary.asp?File=18760&bhcp=1
Network analysis of GWAS data, 2013 Current Opinion in Genetics and Development
Mark DM Leiserson1,2, Jonathan V Eldridge1,2, Sohini Ramachandran2,3 and Benjamin J Raphael1,2
MIF Parsers
The human protein interaction network data can be found from http://www.hprd.org/download
Surendranath V, Niranjan V, Muthusamy B, Gandhi TKB, GronborgM,
Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP,
Ramya MA, Zhao ZX, Chandrika KN, Padma N, Harsha HC et al
(2003) Development of human protein reference database as an initial
platform for approaching systems biology in humans. Genome Res 13:
2363–237
Gerstein lab's human networks
human multinet
http://homes.gersteinlab.org/Khurana-PLoSCompBio-2013/
http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1002886#s4
http://encodenets.gersteinlab.org/
dbCline
UChicago
peptieAtlas
http://www.nature.com/embor/journal/v9/n5/pdf/embor200856.pdf
ensemble
human twin aging expression
http://genomebiology.com/2013/14/7/R75?utm_campaign=10_12_13_genomebiol_Article_Mailing_Reg&utm_content=7387379393&utm_medium=BMCemail&utm_source=Emailvision
http://hongqinlab.blogspot.com/2013/10/mutation-tolerance-in-human-genes-data.html
http://hongqinlab.blogspot.com/2013/03/candidate-rojects-for-r-based-data.html
expression, aging, human, kidny,
http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0020427
9. Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A et al.: Human Protein Reference Database- 2009 update. [Internet]. Nucleic Acids Res 2009, 37: D767-D772.
10. Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M: BioGRID: a general repository for interaction datasets. [Internet]. Nucleic Acids Res 2006, 34:D535-D539.
11. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C et al.: STRING v9.1: protein–protein interaction networks, with increased coverage and integration. [Internet]. Nucleic Acids Res 2013, 41:D808-D815.
12. Razick S, Magklaras G, Donaldson IM: iRefIndex: a consolidated protein interaction database with provenance. [Internet]. BMC Bioinformatics 2008, 9:405.
13. Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B et al.: Reactome: a database of reactions, pathways and biological processes. [Internet]. Nucleic Acids Research 2011, 39:D691-D697.
14. Ewing RM et al.: Large-scale mapping of human protein– protein interactions by mass spectrometry. Molecular Systems Biology 2007, 3:89.
15. Hutchins JRa et al.: Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010, 328:593-599.
16. Rual J-F et al.: Towards a proteome-scale map of the human protein–protein interaction network. Nature 2005, 437:1173-1178.
17. Stelzl U et al.: A human protein–protein interaction network: a resource for annotating the proteome. Cell 2005, 122:957-968.
18. Yu H et al.: Next-generation sequencing to generate interactome datasets. Nat Methods 2011, 8:478-480. 608 Genetics of system biology Current Opinion
From Gilman 2011 Neuron, netbag on autism
Downloaded Information
The data described in previous sections was downloaded from the following public
resources:
GeneOntology annotations from NCBI (01/2009 – ftp://ftp.ncbi.nlm.nih.gov/gene/)
Pathways and enzyme codes from Kyoto Encyclopedia of Genes and Genomes (KEGG)
database (01/2009 – ftp://ftp.genome.jp/pub/kegg/genes/organisms/hsa/)
Domains from InterPro database (01/2009 – ftp://ftp.ebi.ac.uk/pub/databases/interpro/)
Tissue indicators from the TiGER database (09/2009 – http://bioinfo.wilmer.jhu.edu/tiger/)
Protein-protein Interactions:
o BIND with protein complexes (08/2009 – http://bond.unleashedinformatics.com/)
o BioGRID interactions (08/2009 – http://www.thebiogrid.org/downloads.php)
o DIP interactions (10/2009 – http://dip.doe-mbi.ucla.edu/dip/)
o HPRD interactions (10/2009 – http://www.hprd.org/)
o InNetDB interactions (05/2009 – http://hanlab.genetics.ac.cn/sys/intnetdb)
o IntAct (10/2009 – http://www.ebi.ac.uk/intact/)
o BiGG metabolic interactions (04/2009 – http://gcrg.ucsd.edu/Downloads )
o MINT (05/2009 – http://mint.bio.uniroma2.it/mint/)
o MIPS (05/2009 – http://mips.gsf.de/proj/ppi/)
gene expression between human and mouse, PNAS, mike snyder lab, Standford
http://www.pnas.org/content/111/48/17224.full
Labels:
***,
data resources,
genomics,
human,
human network,
networks,
qin,
star
SIAM student chapter log, 2014-2015
For annual report
http://www.siam.org/students/chapters/funding.php
Student membership
http://www.siam.org/students/chapters/free_mem.php
##http://www.siam.org/membership/individual/free.php (This is not a right link for student chapter)
Normally, $500 request is due by Oct 1 for 2014-2015 academic year.
http://www.siam.org/students/chapters/forms.php
Planned activities:
Visit of female mathematician
Latex workshop
R workshop
MATLAB workshop
Student benefits
http://www.siam.org/students/chapters/free_mem.php
Graduate can renew freely for 6 years.
Contacts:
Susan Whitehouse, CAE
Membership Manager
Society for Industrial and Applied Mathematics (SIAM)
3600 Market Street, 6th floor
Philadelphia, PA 19104-2688 USA
Phone: +1 (215) 382-9800, ext 381, 1-800-447-SIAM (toll free US/Canada)
Fax: +1 (215) 386-7999
E-mail: whitehouse@siam.org
Nancy Snell
Membership Coordinator
Society for Industrial & Applied Mathematics (SIAM)
3600 Market Street, 6th floor
Philadelphia, PA 19104-2688 USA
215 382 9800 x 320
nes@siam.org
http://www.siam.org/students/chapters/funding.php
Student membership
http://www.siam.org/students/chapters/free_mem.php
##http://www.siam.org/membership/individual/free.php (This is not a right link for student chapter)
Normally, $500 request is due by Oct 1 for 2014-2015 academic year.
http://www.siam.org/students/chapters/forms.php
Planned activities:
Visit of female mathematician
Latex workshop
R workshop
MATLAB workshop
Student benefits
http://www.siam.org/students/chapters/free_mem.php
Graduate can renew freely for 6 years.
Contacts:
Susan Whitehouse, CAE
Membership Manager
Society for Industrial and Applied Mathematics (SIAM)
3600 Market Street, 6th floor
Philadelphia, PA 19104-2688 USA
Phone: +1 (215) 382-9800, ext 381, 1-800-447-SIAM (toll free US/Canada)
Fax: +1 (215) 386-7999
E-mail: whitehouse@siam.org
Nancy Snell
Membership Coordinator
Society for Industrial & Applied Mathematics (SIAM)
3600 Market Street, 6th floor
Philadelphia, PA 19104-2688 USA
215 382 9800 x 320
nes@siam.org
bio233 review for exam 1
0.5 hr: Let students using Excel to do scatter plot, trend line analysis. MIN, MAX, AVERAGE function usage.
0.5hr: Led students worked on 1 glucose in glycolysis and CAC, how many protons can it pump through the electron transfer chain:
10 NADH and 2 FADH = 10*10+2*6= 112 protons.
20 minutes: Scale of microbe
10/3 proton for 1 ATP in ATP synthase
3.33 proton for 1 ATP
http://biology.stackexchange.com/questions/3186/given-atp-synthases-structure-how-can-3-33-protons-ultimately-synthesize-one-a
http://biology.stackexchange.com/questions/3186/given-atp-synthases-structure-how-can-3-33-protons-ultimately-synthesize-one-a
Friday, September 12, 2014
bio386 reading Stewart 2006 E coli aging paper
Stewart 2005 Plos E coli aging paper.
I asked the student to put red mark all over the paper.
Somehow, "susceptible to aging" was misinterpreted as "not able to aging" by the student.
I asked the student to put red mark all over the paper.
Somehow, "susceptible to aging" was misinterpreted as "not able to aging" by the student.
Thursday, September 11, 2014
bio233, E coli genomes and gene numbers
From
http://www.ncbi.nlm.nih.gov/genome/genomes/167?
I copy-paste and convert this page into Excel.
GC% of the last genome E coli K12 is zero, an obvious mistake.
Q:
Plot genome size with gene number, protein-gene number.
http://www.ncbi.nlm.nih.gov/genome/genomes/167?
I copy-paste and convert this page into Excel.
GC% of the last genome E coli K12 is zero, an obvious mistake.
Q:
Plot genome size with gene number, protein-gene number.
bio233 streak exercise, exam 1 exercises
30 minutes. I use salt and petri dish for students to practice streaking bacterial cells. The salt bounce on petri dish, so it did not work well. I then tried glucose (chemical grade), and it bounce just like salt.
I then spend the rest class working on previous exams. I went through only 4 questions in the spring 2014 exam 1.
One student made a good point that substrate is what an enzyme consumes. So, I should state "product" of an enzyme in the problem stet.
I then spend the rest class working on previous exams. I went through only 4 questions in the spring 2014 exam 1.
One student made a good point that substrate is what an enzyme consumes. So, I should state "product" of an enzyme in the problem stet.
FYE reading 20140911
First, Use monotillation of traxoline as exercise (an elementary school reading exercise)
http://www.bisd.us/gonzalez/STAAR/Reading%20Strand%203rd%20-%205th%20TOT/Handouts/The%20Monotillation%20of%20Traxoline.pdf
context clues
verb => definitions
matching question words with text words
pattern seeking
=> how to improve understanding?
note taking, highlights, translating
Second: microbial mat reading exercise, re-phrasing activities
http://microbes.arc.nasa.gov/STEP/documents/STEP/Microbes@NASA%20Inside.pdf
middle school lesson plan on reading
http://microbes.arc.nasa.gov/download/pdf/Greek_to_Me.pdf
break down, not 'cramming'
In science, figures says a thousand words. So, understanding the figures is the key.
Wednesday, September 10, 2014
bio386 20140910
I used the Feynman lecture on science to explain how science works.
HHMI letter of intent is due next Friday. There is only 1 more class meet to work on the letter.
Students registered for the HHMI writing workshop.
We missed the ABCRMS abstract submission deadline (Sep 5).
Reading assignment on 2006 Plos Biology E coli aging paper. I used Ben Carlson's mom story, and ask student to put red marks all over the paper.
HHMI letter of intent is due next Friday. There is only 1 more class meet to work on the letter.
Students registered for the HHMI writing workshop.
We missed the ABCRMS abstract submission deadline (Sep 5).
Reading assignment on 2006 Plos Biology E coli aging paper. I used Ben Carlson's mom story, and ask student to put red marks all over the paper.
Tuesday, September 9, 2014
IUSE funded projects
quantitative biology, IUSE (nimbios)
http://www.nimbios.org/education/QUBES_2014
https://www.collectiveip.com/grants/NSF:1346584
Interactive Video Vignettes (IVVs)
http://www.nsf.gov/awardsearch/showAward?AWD_ID=1432286&HistoricalAwards=false
Data computing for all
http://www.nsf.gov/awardsearch/showAward?AWD_ID=1432438&HistoricalAwards=false
| Investigator(s): | Christo Dichev dichevc@wssu.edu (Principal Investigator) Darina Dicheva (Co-Principal Investigator) |
| Sponsor: | Winston-Salem State University 601 Martin Luther King Jr., Driv Winston-Salem, NC 27110-0001 (336)750-2413 |
bio233, thursday 20140911, exercises and prepare for exams
todo: print spring 2014 exam and distribute to students.
Negative feedback loop but the class actually ended on 3:40pm.
playdough on activation barrier +/- enzyme Negative feedback loops and fluctuations
Role of NAD+ and NADH in cells, aging, mitochondria and nuclear communication? link to aging?
Negative feedback loop but the class actually ended on 3:40pm.
playdough on activation barrier +/- enzyme Negative feedback loops and fluctuations
Role of NAD+ and NADH in cells, aging, mitochondria and nuclear communication? link to aging?
Moodle grade report
Facility index
Sometimes the "facility index" does not make sense.
This is the average score on the item, expressed as a percentage:
This is the average score on the item, expressed as a percentage:
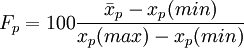 .
.
The higher the facility index, the easier the question is (for this cohort of students).
Discrimination index
This is the product moment correlation coefficient between 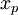 and
and 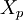 , expressed on a percentage scale. That is,
, expressed on a percentage scale. That is,
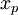 and
and 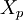 , expressed on a percentage scale. That is,
, expressed on a percentage scale. That is,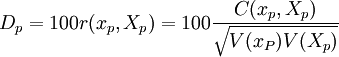 .
.
Subscribe to:
Posts (Atom)





